Users
Social media
- More details here...
- Address
Parc Científic de la Universitat de València C/
Catedrático Agustín Escardino, 9
46980 Paterna (Valencia) Spain - Email:
iu.i2sysbio@uv.es - Phone:
(+34) 963544810
- Address
Links
AI developed to read the “code” of bacterial viruses and design personalised phage therapies
Investigation
AI developed to read the “code” of bacterial viruses and design personalised phage therapies
A research team from the Institute of Integrative Systems Biology (I²SysBio), a joint centre of the University of Valencia and the CSIC, has developed an innovative artificial intelligence system to predict which bacteria can be targeted by bacterial viruses (phages) based on the sequence of a key enzyme: depolymerase. The study has been published in the journal Nature Communications.
Antibiotic resistance is making it increasingly difficult to treat bacterial infections. Phages, which infect bacteria, are emerging as an alternative to conventional antibiotic treatment. However, identifying which phage is effective against each bacterium is complex. This study, led by Robby Concha-Eloko, Beatriz Beamud, Pilar Domingo-Calap and Rafael Sanjuán, proposes the use of artificial intelligence to facilitate this prediction process.
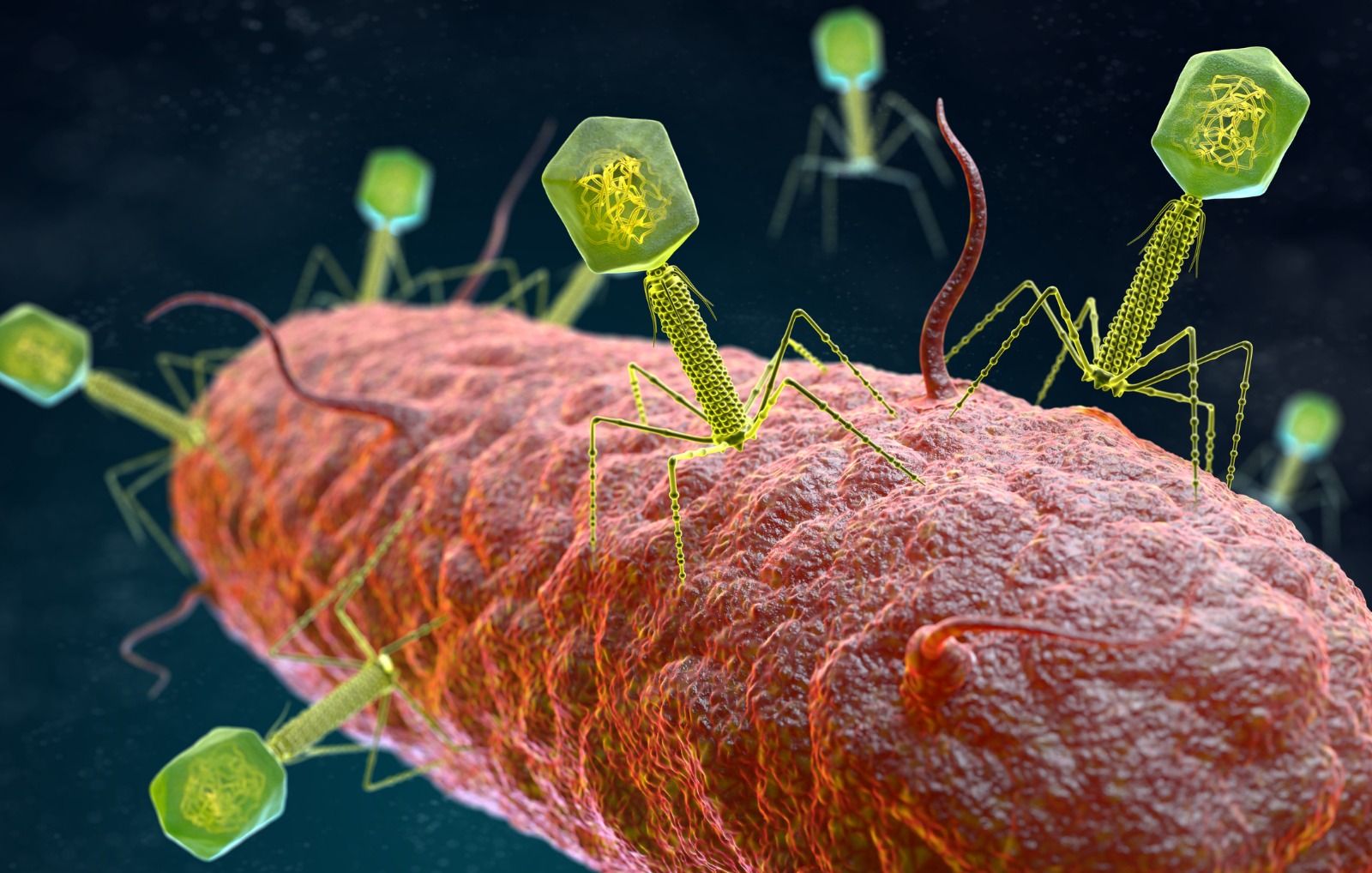
To develop the model, they used the bacterium Klebsiella, listed by the WHO as a priority bacterial pathogen, which is responsible for serious hospital infections and is highly resistant to antibiotics. Klebsiella bacteria are protected by polysaccharide capsules that prevent the activity of antibiotics and the entry of phages. To overcome this barrier, many phages produce depolymerases, enzymes that degrade these capsules and allow the bacteriophage to enter, infect the bacteria and aid in treatment.
However, the enormous genetic diversity of these capsules—more than 100 serotypes of these structures have been recorded in Klebsiella—has made it difficult to predict which phage may be the right one to penetrate the capsule and infect the bacterium. Consequently, this wide variety of capsule serotypes makes Klebsiella an ideal model for studying the interaction between phages and capsules.
A pioneering tool for depolymerase sequencing
With this goal in mind, the research team has developed a pioneering tool that harnesses the genetic information of thousands of Klebsiella bacteria and their “dormant” viruses (prophages) integrated into their genome. By analysing more than 74,000 prophages and nearly 20,000 depolymerase sequences, the researchers have created an unparalleled database that associates each enzyme with the type of bacterial capsule it can degrade.
Using advanced machine learning techniques and artificial intelligence models inspired by natural language processing (similar to those used in machine translation), they have been able to predict with high accuracy the ‘tropism’ or specificity of each depolymerase, i.e., the types of bacterial capsules it can recognise and destroy.
A potential solution against biofilms
This study represents a key advance for biotechnology based on phages or their components, as it allows their specificity to be predicted. This is essential for designing future applications, such as solutions against biofilm, the protective structure that some bacteria form to adhere to surfaces and resist treatment.
Biofilms are increasingly recognised as a significant obstacle in the treatment of infections. In fact, they have been shown to be involved in the chronicity of diseases such as cystic fibrosis, chronic wounds, prosthesis-related infections and urinary tract infections.
‘The use of depolymerase, either in combination with current treatments (antibiotics or antimicrobial peptides) or potentially as an immune system enhancer, can address issues related to biofilm formation, reducing the risk of treatment failure,’ explains Robby-Concha.
‘Compared to the traditional method, which is based on a tedious process of searching for and testing phages to find an effective depolymerase, artificial intelligence models allow us to predict their specificity in silico,’ says the researcher. In this regard, the method demonstrated in the study allows the generation of depolymerase libraries that can be used to extract the most effective enzyme, optimising the degradation of the capsule and, subsequently, the biofilm. This represents a breakthrough in the development of anti-biofilm strategies as viable candidates for future clinical studies.
New applications targeting priority pathogens
According to Robby-Concha, one of the developers of this pioneering tool, although Klebsiella has been used as a model, this methodology can be used against any other capsule-producing bacterium. This includes most of the priority pathogens listed by the WHO.
In summary, this study proposes resolving predictions of interaction between phages and hosts in two ways. First, by leveraging data contained in bacterial genomes (prophages), which provides important training data; and second, by proposing an architecture that allows the model to be trained with all bacterial species at once (in an integrative manner).
Concha-Eloko, R., Beamud, B., Domingo-Calap, P. et al. Unlocking data in Klebsiella lysogens to predict capsular type-specificity of phage depolymerases. Nat Commun 16, 8798 (2025). https://doi.org/10.1038/s41467-025-63861-w