Users
Social media
- More details here...
- Address
Parc Científic de la Universitat de València C/
Catedrático Agustín Escardino, 9
46980 Paterna (Valencia) Spain - Email:
iu.i2sysbio@uv.es - Phone:
(+34) 963544810
- Address
Links
Specialists from I2SysBio, CSIC and FISABIO analyze the genetic keys to the virulence of tuberculosis
Investigation & Education
Thesis
Specialists from I2SysBio, CSIC and FISABIO analyze the genetic keys to the virulence of tuberculosis

Research staff from the Institute of Biomedicine of Valencia (CSIC), the Institute of Integrative Systems Biology (CSIC-UV) and the Foundation for the Promotion of Health and Biomedical Research of the Generalitat Valenciana have analyzed the genomic determinants of speciation and the spread of tuberculosis. The work, published in the journal Science Advances, identifies the phoR gene as the key to a system involved in the virulence of tuberculosis and expands knowledge about the evolution of the bacteria that cause this pathology in animals and humans.
Tuberculosis is an infectious disease caused by the bacteria Mycobacterium tuberculosis, which causes devastating mortality in humans and animals, and also entails significant economic losses. Knowing how the different bacterial lineages differ increases the understanding of the origins of the bacteria that cause the disease and the genetic mechanisms involved.
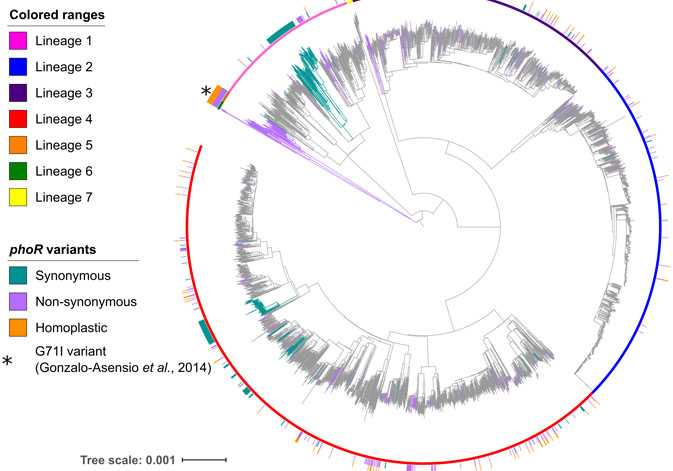
The researcher at the Institute of Biomedicine of Valencia (IBV, CSIC) Iñaki Comas and the researcher at the Mixed Unit FISABIO/Universitat de València-I2SysBio Álvaro Chiner (currently, member of the IBV, CSIC) explain that “for To understand the population genomic events that led to the appearance of the tuberculosis pathogen, we have worked with the Mycobacterium tuberculosis Complex or MTBC, which comprises a group of mycobacteria made up of Mycobacterium tuberculosis and Mycobacterium africanum, which affect humans, as well as a series of pathogens isolated from other mammalian species known as Mycobacterium bovis, Mycobacterium pinnipedii, Mycobacterium orygis and Mycobacterium microti, among others.”
The increasing availability of population genomic data has allowed a better understanding of the genotypic and ecological differentiation between closely related bacteria. This has allowed us to develop theoretical models of how bacterial species and the genomic regions involved emerge.
“This study applies these models for the first time to a pathogen that affects humans. For this purpose, the bacteria most closely related to MTBC have been studied, known as Mycobacterium canettii or MCAN, a strain isolated from the Horn of Africa. Our analysis confirms the hypothesis that both shared a common genetic pool. In addition, we have taken advantage of the availability of sequences "genomic analyzes of thousands of clinical strains of the MTBC complex, as well as close relatives such as MCAN, to identify new genomic determinants in the appearance and subsequent spread of MTBC," adds Iñaki Comas.
The researchers have identified the phoR gene as the key to a system involved in virulence, and that played a fundamental role in the evolution of the Mycobacterium tuberculosis Complex. "Previous work had shown that phoR mutations played a central role in the adaptation of the pathogen to different host species. We have demonstrated the link of the phoR gene with the early spread of human tuberculosis, as well as in later expansions. Our work also demonstrates that the study of the evolution of pathogens helps to understand the determinants of their past and present virulence," concludes Comas.
Researchers from the CIBER of Epidemiology and Public Health of Valencia, the University of Oslo, the University of Helsinki, the Swiss Tropical and Public Health Institute of Switzerland, the University of Basel, the Microbiotica BioData Innovation Center of the United Kingdom, and the Francis Crick Institute of the United Kingdom.
Article: Á. Chiner-Oms, L. Sánchez-Busó, J. Corander, S. Gagneux, S. Harris, D. Young, F. GonzálezCandelas, I. Comas. "Genomic determinants of speciation and spread of the Mycobacterium tuberculosis complex." Science Advances. DOI: 10.1126/sciadv.aaw3307